Tempus xF

xF Liquid Biopsy Test
Features
- 1
SNVs (single nucleotide variants) & insertions and deletions (indels): 105 Genes
실행 가능한 돌연변이에 초점을 맞춘 DNA sequencing
- 2
Immunotherapy Metrics: Microsatellite Instability High (MSI-H)
- 3
Copy Number Gains/Amplifications: 6 Genes
- 4
Copy Number Losses/Deletions: 2 Genes (BRCA1 & BRCA2)
- 5
Rearrangements/Fusion: 7 Genes
- 6
DNA Sequencing Depth: average 20,000x (raw reads)/5,000x (unique reads)
- 7
폐 선암종 또는 기타 고형종양 (뇌암, 육종 제외) 진단 시 검체가 불충분한 경우 권장
Specimen types
- 1
고형 종양 조직 용 말초 혈액: 8.5ml streck tube 2개
xF Liquid Biopsy로의 자동 변환 옵션
xT test시 조직의 퀄리티가 낮거나 검체량이 부족한
경우 xF test로 자동 변환
Request form 작성시 선택:
- xF로 즉시변환
- xF 변환 전에 추가 조직 요청
To monitor mechanisms of resistance
최근 발표된 데이터에서 아래의 진행성 암 환자에 대한 first line 표적 치료 시 발생하는 resistant mechanism 확인.

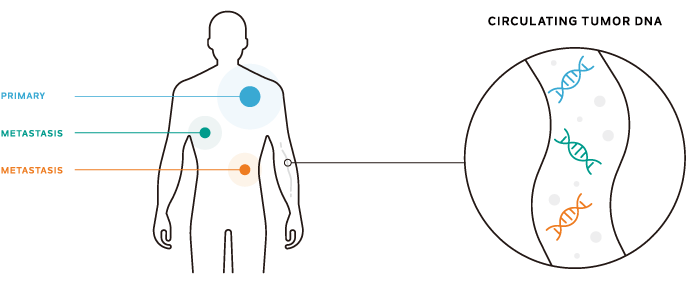
To gain a comprehensive view of a patient’s metastatic disease
Inter-, intra-tumor의 이질성은 고형 조직 및 cfDNA 액체 생검 테스트에 활용 시 중첩되지 않고 잠재적으로 actionable한 결과 도출. 이러한 접근은 모든 종양 부위에 걸쳐 포괄적인 평가를 제공할 수 있음(8)(9)(10)
REFERENCES
- Chabon, J., Simmons, A., Lovejoy, A. et al. (2016). doi:10.1038/ncomms11815
- Ihuegbu, N., Banks, K., Fairclough, S. et al. (2016). doi:10.1200/JCO.2016.34.15_suppl.e20643
- Fribbens, C., O’Leary, B., Kilburn, L. et al. (2016). doi:10.1200/JCO.2016.67.3061
- Weigelt, B., Comino-Méndez, I., de Bruijn, I. et al. (2017). doi:10.1158/1078-0432.CCR-17-0544
- Azad, A., Volik, S., Wyatt, A. et al. (2015). doi:10.1158/1078-0432.CCR-14-2666
- Parikh, A.R., Leshchiner, I., Elagina, L. et al. (2019). doi:10.1038/s41591-019-0561-9
- Clifton, K., Rich, T., Parseghian, C. et al. (2019). doi:10.1200/PO.19.00141
- Aggarwal, C., Thompson, J., Black, T., et al. (2019). doi:10.1001/jamaoncol.2018.4305
- Schwaederle, M., Patel, S., Husain, H. et al. (2017). doi:10.1158/1078-0432.CCR-16-2497
- Maxwell, K., Soucier-Ernst, D., Tahirovic, E. et al. (2017). doi:10.1007/s10549-017-4257-x
Product

LIQUID BIOPSY
xF Plasma ctDNA (105 genes)Auto-conversion options from xT in event of tissue NGS failure/QNS
Testing for Solid Tumors
| CANCER SITE | PANEL TEST | Normal specimen from clinic | Cancer specimen from clinic | Cancer specimen | |
|---|---|---|---|---|---|
| Solid tumor where a biopsy specimen is available | xT Solid Tumor | - | - | FFPE resection / biopsy | |
| Solid tumor where a biopsy specimen is not readily available | xF Liquid Biopsy | - | Peripheral blood | - |
Testing for Hematologic Malignancies
| CANCER SITE | PANEL TEST | Normal specimen from clinic | Cancer specimen from clinic | Cancer specimen | |
|---|---|---|---|---|---|
| Lymphoma (not in leukemic form) | xT Solid Tumor | Normal saliva | - | FFPE resection / biopsy | |
| Leukemia, MDS, MPN | xT Heme | - | Peripheral blood or bone marrow aspirate | OR | FFPE bone marrow clot |
| Multiple myeloma | xT Heme | Normal saliva | bone marrow aspirate | OR | FFPE bone marrow clot |
xF GENE PANEL
A non-invasive, liquid biopsy panel of 105 genes focused on oncogenic and resistance mutations in cell-free DNA (cfDNA). This panel is designed to provide clinical decision support for solid tumors.
- SNVs (single nucleotide variants) and insertions and deletions (indels) are detected in all 105 genes
- Copy Number Amplifications (CNAs), Copy Number Deletions (CNDs)¹, and gene rearrangements (translocations) are detected in a subset of genes
- DNA Sequencing Depth: average 20,000x (raw reads)/5,000x (unique reads)
- Specimen Requirements: Two Streck tubes of peripheral blood (8.5mL each)
The report includes genomic alterations in select genes, microsatellite instability status², median variant allele fraction (mVAF), therapy options and clinical trials matched to the patient’s genomic profile, as well as clinical history.
xF GENE PANEL
| AKT1 | BRAF | CDK6 | FGFR1 | HRAS | MAP2K1 | MYCN | PDGFRA | RET | TERT |
| AKT2 | BRCA1 | CDKN2A | FGFR2 | IDH1 | MAP2K2 | NF1 | PDGFRB | RHEB | TP53 |
| ALK | BRCA2 | CTNNB1 | FGFR3 | IDH2 | MAPK1 | NF2 | PIK3CA | RHOA | TSC1 |
| APC | BTK | DDR2 | FGFR4 | JAK1 | MET | NFE2L2 | PIK3R1 | RIT1 | TSC2 |
| AR | CCND1 | DPYD | FLT3 | JAK2 | MLH1 | NOTCH1 | PMS2 | RNF43 | UGT1A1 |
| ARAF | CCND2 | EGFR | FOXL2 | JAK3 | MPL | NPM1 | PTCH1 | ROS1 | VHL |
| ARID1A | CCND3 | ERBB2 (HER2) | GATA3 | KDR | MSH2 | NRAS | PTEN | SDHA | |
| ATM | CCNE1 | ERRFI1 | GNA11 | KEAP1 | MSH3 | NTRK1 | PTPN11 | SMAD4 | |
| ATR | CD274 (PD-L1) | ESR1 | GNAQ | KIT | MSH6 | PALB2 | RAD51C | SMO | |
| B2M | CDH1 | EZH2 | GNAS | KMT2A | MTOR | PBRM1 | RAF1 | SPOP | |
| BAP1 | CDK4 | FBXW7 | HNF1A | KRAS | MYC | PDCD1LG2 | RB1 | STK11 |
GENE REARRANGEMENTS
| ALK | BRAF | FGFR2 | FGFR3 | NTRK1 | RET | ROS1 |
COPY NUMBER GAINS
| CCNE1 | CD274 (PD-L1) | EGFR | ERBB2 (HER2) | MET | MYC |
COPY NUMBER LOSSES
| BRCA1 | BRCA2 |
PERFORMANCE SPECIFICATIONS
| Variant Allele Fraction (VAF) | Sensitivity3 | |
|---|---|---|
| Single Nucleotide Variants (SNV) | >0.5% | >99.9% |
| 0.50% | >99.9% | |
| 0.25% | 97% | |
| 0.10%4 | 70.4% | |
| Insertions and Deletions | >0.5% | 98.8% |
| 0.50% | 96.0% | |
| 0.25% | 81.0% | |
| Copy Number Amplifications (CNAs) | >0.5% | >99.9% |
| 0.5% | >99.9% | |
| Rearrangements/Fusions | >0.5% | 97.4% |
| 0.50% | 70.8% |
ANALYTICAL SPECIFICITY
| Variant Type | Specificity3 |
|---|---|
| SNV | >99.9% |
| INDEL | >99.9% |
| CNA | 96.2% |
| Fusion | >99.9% |
1. BRCA1 and BRCA2 copy number loss are reported when detected
2. MSI status will be reported when the specimen is determined to be MSI-High
3. Established using reference materials
4. For selected hot spot regions