Tempus xT

Discover, design, and
deliver the right drug for
the right patient
Tempus provides integrated solutions to help meet your needs from early discovery through commercialization.
최신 genomic sequencing 및 최첨단 기술을 사용하여 up-to-date 치료 옵션을 제안하며 치료로 직결될 수 있는 insight를 제공합니다.
xT Solid Tumor + Normal Test
Features
- 1
648개 유전자 패널
실행 가능한 돌연변이에 초점을 맞춘 DNA sequencing
- 2
종양+정상 일치 (혈액 또는 타액) 분석
- 65개 incidental germline 포함
- 종양 단독 분석보다 위양성율이 27.7% 낮음
- 3
MSI status, Tumor Mutational Burden (TMB)
- 4
IHC 염색 (선택사항)
- PD-L1 (22C3, SP142, 28-8 clones)
- MMR
- 5
RNA sequencing을 통한 전체 전사체 분석
Specimen types
- 1
고형암조직 (예) 폐, 대장, 유방, 췌장 등
- 2
정상 (혈액 또는 타액): 8.5ml streck tube 2개
xT Solid Tumor Only Test
Features
- 1
648개 유전자 패널
실행 가능한 돌연변이에 초점을 맞춘 DNA sequencing
- 2
MSI status, Tumor Mutational Burden (TMB)
- 3
IHC 염색 (선택사항)
- PD-L1 (22C3, SP142, 28-8 clones)
- MMR
- 4
RNA sequencing을 통한 전체 전사체 분석
Specimen types
- 1
고형암조직 (예) 폐, 대장, 유방, 췌장 등
xT Hematologic Test
Features
- 1
648개 유전자 패널
실행 가능한 돌연변이에 초점을 맞춘 DNA sequencing
- 2
종양+정상 일치 (혈액 또는 타액) 분석
- 65개 incidental germline 포함
- 종양 단독 분석보다 위양성율이 27.7% 낮음
- 3
MSI status, Tumor Mutational Burden (TMB)
- 4
IHC 염색 (선택사항)
- PD-L1 (22C3, SP142, 28-8 clones)
- MMR
- 5
RNA sequencing을 통한 전체 전사체 분석
Specimen types
- 1
림프종: 림프절 생검 또는 골수 흡인, 응고체 또는 생검; 혈액 내 순환 림프종 침범이 충분할 경우 말초 혈액 허용
- 정상 검체 매칭 시 타액 필요 (정상 매칭을 위한 혈액은 허용되지 않음)
- 2
백혈병: 말초 혈액 또는 골수 흡인, 응고체 또는 생검
– 정상 매칭은 허용되지 않음
- 3
골수종: 골수 흡인, 응고체 또는 생검
– 정상 검체 매칭 시 타액 필요 (정상 매칭을 위한 혈액은 허용되지 않음)
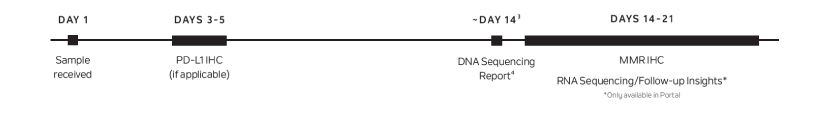
Result Timing and Delivery Method
- Reports are delivered in approximately two weeks from the time Tempus has received both the tumor and normal samples (if applicable).
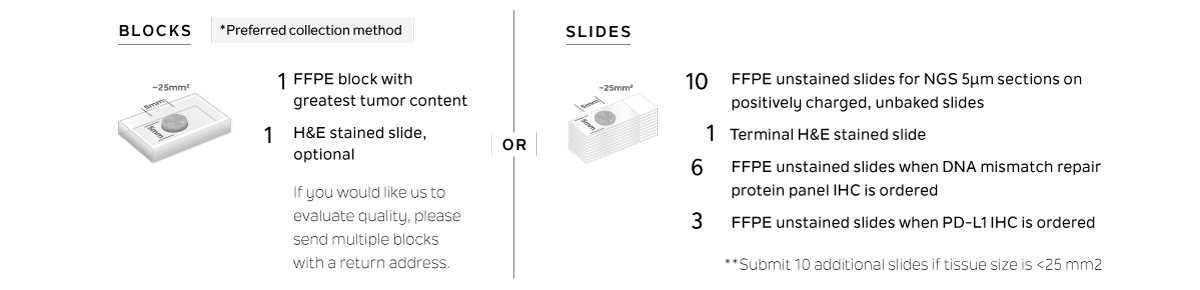
TISSUE REQUIREMENTS
- 2개의 식별자로 라벨이 붙은 환자 샘플
- 종양 샘플은 가능한 최근에 채취
- FFPE 고정 요건
- 6~72시간 동안 10% 포르말린 고정 (neutral buffered), paraffin embedded
- 샘플의 석회질 제거는 바람직하지 않음 (EDTA 허용).
- 종양은 샘플의 20% 이상 (ratio of tumor nuclei to benign nuclei)
- 최적 종양 크기 = 25 mm², 최소 = 5 mm²
Product

TISSUE
xT Solid Tumor + Heme NGS (648 genes)DNA + RNA transcriptome validated fusion detection
Immune Biomarkers -TMB, MSI, PD-L1 (IHC)
HRD & TO add-on tests available
Incidental Germline (Tumor-Normal Match)
DPYD & UGT1A1 add-on tests available (Tumor-Normal Match
LIQUID BIOPSY
xF Plasma ctDNA (105 genes)Auto-conversion options from xT in event of tissue NGS failure/QNS
Testing for Solid Tumors
| CANCER SITE | PANEL TEST | Normal specimen from clinic | Cancer specimen from clinic | Cancer specimen | |
|---|---|---|---|---|---|
| Solid tumor where a biopsy specimen is available | xT Solid Tumor | - | - | FFPE resection / biopsy | |
| Solid tumor where a biopsy specimen is not readily available | xF Liquid Biopsy | - | Peripheral blood | - |
Testing for Hematologic Malignancies
| CANCER SITE | PANEL TEST | Normal specimen from clinic | Cancer specimen from clinic | Cancer specimen | |
|---|---|---|---|---|---|
| Lymphoma (not in leukemic form) | xT Solid Tumor | Normal saliva | - | FFPE resection / biopsy | |
| Leukemia, MDS, MPN | xT Heme | - | Peripheral blood or bone marrow aspirate | OR | FFPE bone marrow clot |
| Multiple myeloma | xT Heme | Normal saliva | bone marrow aspirate | OR | FFPE bone marrow clot |
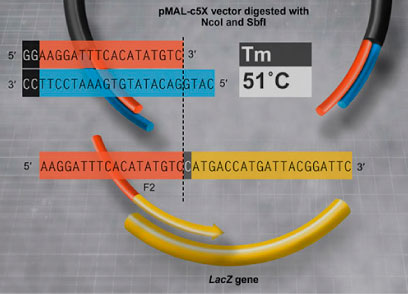
xT GENE PANEL
648 gene panel focused on actionable mutations by DNA sequencing
- Specimen: tumor and matched normal (peripheral blood or saliva)
- Genomic rearrangements are detected on 22 genes by DNA sequencing (others detected by RNA sequencing)
- Microsatellite instability status and tumor mutational burden are included in the xT report
- Average coverage ~ 500x
Full transcriptome by RNA sequencing
- Specimen: tumor or heme
- Unbiased gene rearrangement detection from fusion transcripts are clinically validated
- Whole transcriptome RNA expression counts are analytically validated
- Sequenced at a minimum of 25 million reads, average 50 million reads
For use with xT | 648 GENE PANEL REPORTS
| ABCB1 | BLM** | CHEK2** | EPHB1 | FGFR2 | HLA-DMA | JAK3 | MLH1** | PAX5 | PTPN22 | SLX4 | TNF |
| ABCC3 | BMPR1A** | CIC | EPHB2 | FGFR3 | HLA-DMB | JUN | MLH3 | PAX7 | PTPRD | SMAD2 | TNFAIP3 |
| ABL1 | BRAF | CIITA | EPOR | FGFR4 | HLA-DOA | KAT6A | MLLT3 | PAX8 | PTPRT | SMAD3 | TNFRSF14 |
| ABL2 | BRCA1** | CKS1B | ERBB2 | FH** | HLA-DOB | KDM5A | MN1 | PBRM1 | QKI | SMAD4** | TNFRSF17 |
| ABRAXAS1 | BRCA2** | CREBBP | (HER2) | FHIT | HLA-DPA1 | KDM5C | MPL | PCBP1 | RAC1 | SMARCA1 | TNFRSF9 |
| ABRAXAS1 | BRCA2** | CREBBP | (HER2) | FHIT | HLA-DPA1 | KDM5C | MPL | PCBP1 | RAC1 | SMARCA1 | TNFRSF9 |
| ACTA2 | BRD4 | CRKL | ERBB3 | FLCN** | HLA-DPB1 | KDM5D | MRE11 | PDCD1 | RAD21 | SMARCA4** | TOP1 |
| ACVR1 | BRIP1** | CRLF2 | ERBB4 | FLT1 | HLA-DPB2 | KDM6A | MS4A1 | PDCD1LG2 | RAD50 | SMARCB1** | TOP2A |
| (ALK2) | BTG1 | CSF1R | ERCC1 | FLT3 | HLA-DQA1 | KDR | MSH2** | PDGFRA** | RAD51 | SMARCE1 | TP53** |
| ACVR1B | BTK | CSF3R | ERCC2 | FLT4 | HLA-DQA2 | KEAP1 | MSH3** | PDGFRB | RAD51B | SMC1A | TP63 |
| AGO1 | BUB1B | CTC1 | ERCC3 | FNTB | HLA-DQB1 | KEL | MSH6** | PDK1 | RAD51C** | SMC3 | TPM1 |
| AJUBA | C11orf65 | CTCF | ERCC4 | FOXA1 | HLA-DQB2 | KIF1B | MTAP | PHF6 | RAD51D** | SMO | TPMT |
| AKT1 | C3orf70 | CTLA4 | ERCC5 | FOXL2 | HLA-DRA | KIT** | MTHFD2 | PHGDH | RAD54L | SOCS1 | TRAF3 |
| AKT2 | C8orf34 | CTNNA1 | ERCC6 | FOXO1 | HLA-DRB1 | KLF4 | MTHFR | PHLPP1 | RAF1 | SOD2 | TRAF7 |
| AKT3 | CALR | CTNNB1 | ERG | FOXO3 | HLA-DRB5 | KLHL6 | MTOR | PHLPP2 | RANBP2 | SOX10 | TSC1** |
| ALK | CARD11 | CTRC | ERRFI1 | FOXP1 | HLA-DRB6 | KLLN | MTRR | PHOX2B** | RARA | SOX2 | TSC2** |
| AMER1 | CARM1 | CUL1 | ESR1 | FOXQ1 | HLA-E | KMT2A | MUTYH** | PIAS4 | RASA1 | SOX9 | TSHR |
| APC** | CASP8 | CUL3 | ETS1 | FRS2 | HLA-F | KMT2B | MYB | PIK3C2B | RB1** | SPEN | TUSC3 |
| APLNR | CASR | CUL4A | ETS2 | FUBP1 | HLA-G | KMT2C | MYC | PIK3CA | SPINK1 | TYMS | |
| APOB | CBFB | CUL4B | ETV1 | FUS | HNF1A | KMT2D | MYCL | PIK3CB | RECQL4 | SPOP | U2AF1 |
| AR | CBL | CUX1 | ETV4 | G6PD | HNF1B | KRAS | MYCN | PIK3CD | RET** | SPRED1 | UBE2T |
| ARAF | CBLB | CXCR4 | ETV5 | GABRA6 | HOXA11 | L2HGDH | MYD88 | PIK3CG | RHEB | SRC | UGT1A1 |
| ARHGAP26 | CBLC | CYLD | ETV6** | GALNT12 | HOXB13 | LAG3 | MYH11 | PIK3R1 | RHOA | SRSF2 | UGT1A9 |
| ARHGAP35 | CBR3 | CYP1B1 | EWSR1 | GATA1 | HRAS | LATS1 | NBN** | PIK3R2 | RICTOR | STAG2 | UMPS |
| ARID1A | CCDC6 | CYP2D6 | EZH2 | GATA2** | HSD11B2 | LCK | NCOR1 | PIM1 | RINT1 | STAT3 | VEGFA |
| ARID1B | CCND1 | CYP3A5 | FAM46C | GATA3 | HSD3B1 | LDLR | NCOR2 | RIT1 | STAT4 | VEGFB | |
| ARID2 | CCND2 | CYSLTR2 | FANCA | GATA4 | HSD3B2 | LEF1 | NF1** | PLCG2 | RNF139 | STAT5A | VHL** |
| ARID5B | CCND3 | DAXX | FANCB | GATA6 | HSP90AA1 | LMNA | NF2** | PML | RNF43 | STAT5B | VSIR |
| ASNS | CCNE1 | DDB2 | FANCC | GEN1 | HSPH1 | LMO1 | NFE2L2 | PMS1 | ROS1 | STAT6 | WEE1 |
| ASPSCR1 | CD19 | DDR2 | FANCD2 | GLI1 | IDH1 | LRP1B | NFKBIA | PMS2** | RPL5 | STK11** | WNK1 |
| ASXL1 | CD22 | DDX3X | FANCE | GLI2 | IDH2 | LYN | NHP2 | POLD1** | RPS15 | SUFU** | |
| ATIC | CD274 | DICER1** | FANCF | GNA11 | IDO1 | LZTR1 | NKX2-1 | POLE** | RPS6KB1 | SUZ12 | WRN |
| ATM** | (PD-L1) | DIRC2 | FANCG | GNA13 | IFIT1 | MAD2L2 | NOP10 | POLH | RPTOR | SYK | WT1** |
| ATP7B | CD40 | DIS3 | FANCI | GNAQ | IFIT2 | MAF | NOTCH1 | POLQ | RRM1 | SYNE1 | XPA |
| ATR | CD70 | DIS3L2 | FANCL | GNAS | IFIT3 | MAFB | NOTCH2 | POT1 | RSF1 | TAF1 | XPC |
| ATRX | CD79A | DKC1 | FANCM | GPC3 | IFNAR1 | MAGI2 | NOTCH3 | POU2F2 | RUNX1 ** | TANC1 | XPO1 |
| AURKA | CD79B | DNM2 | FAS | GPS2 | IFNAR2 | MALT1 | NOTCH4 | PPARA | RUNX1T1 | TAP1 | XRCC1 |
| AURKB | CDC73 | DNMT3A | FAT1 | GREM1 | IFNGR1 | MAP2K1 | NPM1 | PPARD | RXRA | TAP2 | XRCC2 |
| AXIN1 | CDH1** | DOT1L | FBXO11 | GRIN2A | IFNGR2 | MAP2K2 | NQO1 | PPARG | SCG5 | TARBP2 | XRCC3 |
| AXIN2** | CDK12 | DPYD | FBXW7 | GRM3 | IFNL3 | MAP2K4 | NRAS | PPM1D | SDHA** | TBC1D12 | YEATS4 |
| AXL | CDK4** | DYNC2H1 | FCGR2A | GSTP1 | IKBKE | MAP3K1 | NRG1 | PPP1R15A | SDHAF2** | TBL1XR1 | ZFHX3 |
| B2M | CDK6 | EBF1 | FCGR3A | H19 | IKZF1 | MAP3K7 | NSD1 | PPP2R1A | SDHB** | TBX3 | ZMYM3 |
| BAP1** | CDK8 | ECT2L | FDPS | H3F3A | IL10RA | MAPK1 | NSD2 | PPP2R2A | SDHC** | TCF3 | ZNF217 |
| BARD1** | CDKN1A | EGF | FGF1 | HAS3 | IL15 | MAX** | NT5C2 | PPP6C | SDHD** | TCF7L2 | ZNF471 |
| BCL10 | CDKN1B | EGFR** | FGF10 | HAVCR2 | IL2RA | MC1R | NTHL1** | PRCC | SEC23B | TCL1A | ZNF620 |
| BCL11B | CDKN1C | EGLN1 | FGF14 | HDAC1 | IL6R | MCL1 | NTRK1 | PRDM1 | SEMA3C | TERT* | ZNF750 |
| BCL2 | CDKN2A** | EIF1AX | FGF2 | HDAC2 | IL7R | MDM2 | NTRK2 | PREX2 | SETBP1 | TET2 | ZNRF3 |
| BCL2L1 | CDKN2B | ELF3 | FGF23 | HDAC4 | ING1 | MDM4 | NTRK3 | PRKAR1A** | SETD2 | TFE3 | ZRSR2 |
| BCL2L11 | CDKN2C | ELOC | FGF3 | HGF | INPP4B | MED12 | NUDT15 | PRKDC | SF3B1 | TFEB | |
| BCL6 | CEBPA** | (TCEB1) | FGF4 | HIF1A | IRF1 | MEF2B | NUP98 | PRKN | SGK1 | TFEC | |
| BCL7A | CEP57 | EMSY | FGF5 | HIST1H1E | IRF2 | MEN1** | OLIG2 | PRSS1 | SH2B3 | TGFBR1 | |
| BCLAF1 | CFTR | ENG | FGF6 | HIST1H3B | IRF4 | MET** | P2RY8 | PTCH1** | SHH | TGFBR2 | |
| BCOR | CHD2 | EP300 | FGF7 | HIST1H4E | IRS2 | MGMT | PAK1 | PTCH2 | SLC26A3 | TIGIT | |
| BCORL1 | CHD4 | EPCAM** | FGF8 | HLA-A | ITPKB | MIB1 | PALB2** | PTEN** | SLC47A2 | TMEM127** | |
| BCR | CHD7 | EPHA2 | FGF9 | HLA-B | JAK1 | MITF | PALLD | PTPN11 | SLC9A3R1 | TMEM173 | |
| BIRC3 | CHEK1 | EPHA7 | FGFR1 | HLA-C | JAK2 | MKI67 | PAX3 | PTPN13 | SLIT2 | TMPRSS2 |
GENE REARRANGEMENTS BY DNA SEQUENCING
| ABL1 | EGFR** | FGFR3 | NTRK2 | PML | TFE3 |
| ALK | ETV6** | MYB | NTRK3 | RARA | TMPRSS2 |
| BCR | EWSR1 | NRG1 | PAX8 | RET | |
| BRAF | FGFR2 | NTRK1 | PDGFRA | ROS1 |
* Includes promoter region
** Genes in which incidental germline findings are reported
In addition to reporting on somatic variants, when a normal sample is provided, Tempus reports
germline incidental findings on a limited set of variants associated with inherited cancer syndromes within genes selected based on recommendations from the American College of Medical Genetics (ACMG), the National Comprehensive Cancer Network (NCCN), and/or published literature. Exons and select intronic regions only. Detailed list provided upon request.