DNA Extraction kit

The Monarch® HMW DNA Extraction Kits provide a rapid and reliable process for extracting high molecular weight DNA (HMW DNA) from biological samples. These kits utilize an optimized process that combines gentle cell lysis with a tunable fragment length generation, followed by precipitation of the extracted DNA onto the surface of large glass beads. DNA size ranges from 50-250 kb for the standard protocol and into the Mb range for certain sample types when the lowest agitation speeds are used. Purified DNA is recovered in high yield with excellent purity, including nearly complete removal of RNA, and is ready for use in downstream applications including long-read sequencing.
Feature
Reasons to Choose the Monarch HMW DNA Extraction Kits
- 1
Extremely fast, user-friendly protocols utilizing a novel glass-bead-based approach
- 2
Reproducibly purify high molecular weight genomic DNA (HMW DNA) from various sample types
- 3
Tune fragment length by varying agitation speeds during lysis; achieve DNA in the Megabase size range with low speeds
- 4
Achieve outstanding results when compared to other commercially available solutions
- 5
Excellent performance in long read sequencing
Extremely fast, user-friendly protocols utilizing a novel glass-bead-based approach
Comparison of HMW DNA Extraction Methods
| Extraction Method | Approximate protocol/workflow time* | Notes** | ||
|---|---|---|---|---|
| Cells | Blood | Tissue | ||
| NEB Monarch HMW DNA Extraction | 30 minutes | 60 minutes | 90 minutes | Fast, convenient, High yield, pure, tunable DNA size |
| Circulomics® Nanobind® | > 60 minutes | 60 minutes | >2 hours | Several hands-on steps, difficult to process some samples due to high viscosity. |
| Phenol/Chloroform Extraction | > 6 hours | > 6 hours | > 6 hours | Hazardous reagents, lengthy workflow, DNA is difficult to dissolve |
| Qiagen® MagAttract® | Not compatible | ~60 minutes | Overnight + 40 minutes |
DNA is heavily sheared |
| Qiagen® Genomic Tips | ~3.5 hours | 3.5 hours | ~4 hours | Long workflow, sub-optimal yields, not very high molecular weight |
| Revolugen® FireMonkey® | ~45 minutes | >60 minutes | Not compatible | Only second eluate is used (very low recovery); DNA heated to 80°C leading to denaturing |
*workflow times are estimated based on protocol protocols published as of July 2020, and internal testing or usage
**Notes are based on internal testing or usage
Workflow for Processing Cell Samples
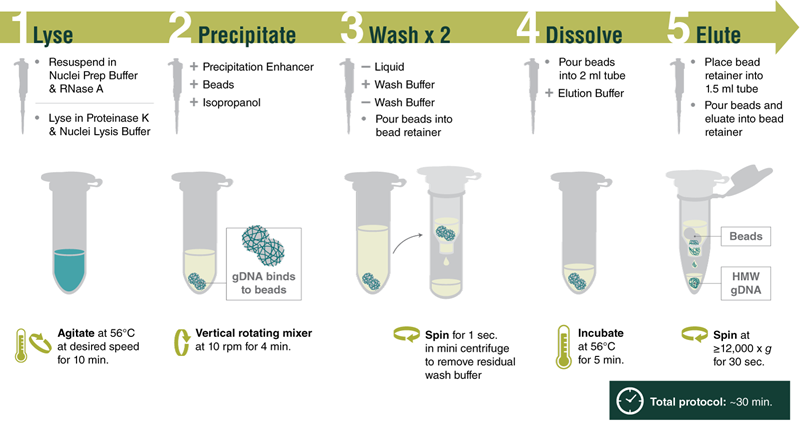
Reproducibly purify high molecular weight genomic DNA (HMW DNA) from various sample types
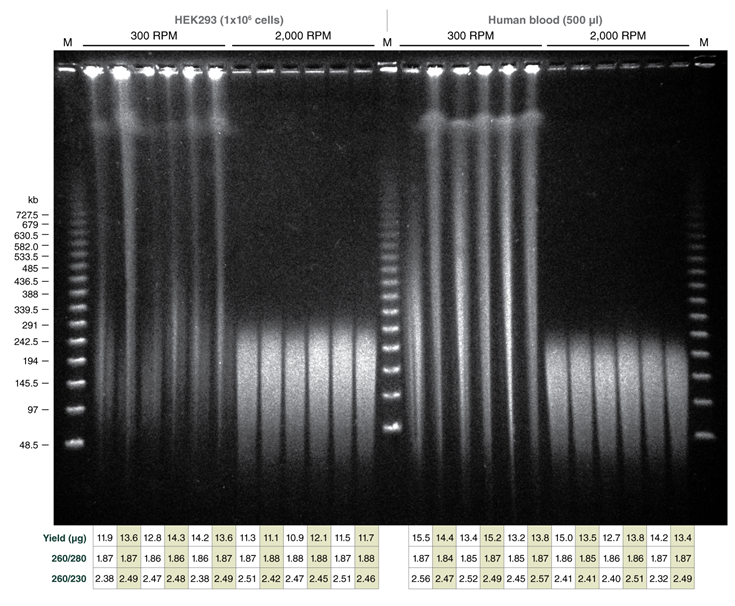
Reproducible Extraction of HMW DNA from Cells and Blood with the Monarch HMW DNA Extraction Kit
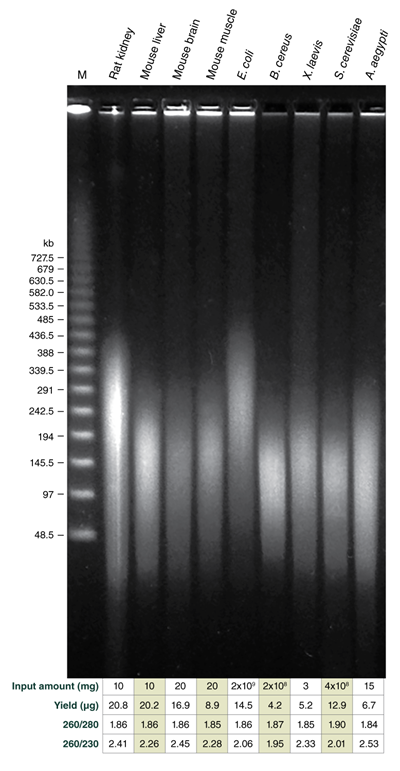
The Monarch HMW DNA Extraction Kit for Tissue efficiently purifies high-quality, HMW DNA from a variety of sample types
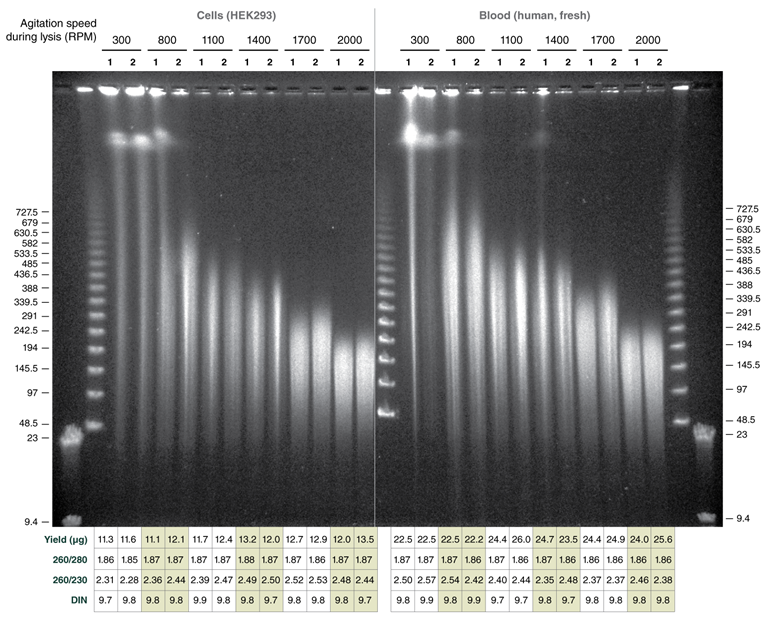
Tune fragment length by varying agitation speeds during lysis; achieve DNA in the Megabase size range with low speeds
Use of varying agitation speeds during lysis produces tunable fragment length of extracted HMW genomic DNA from cells and blood
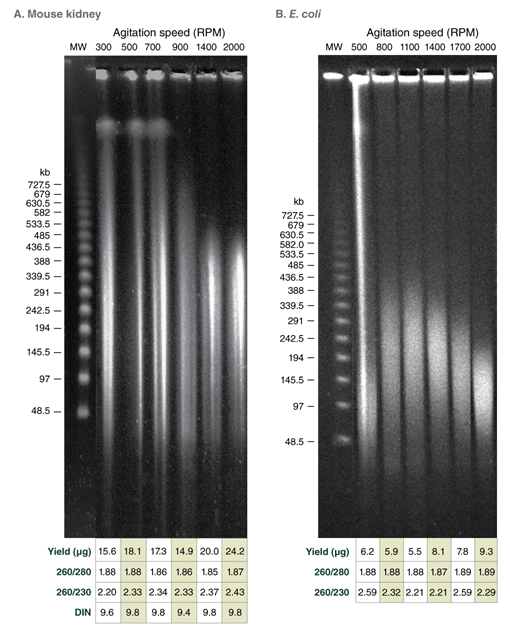
Use of varying agitation speeds during lysis produces tunable fragment length for HMW genomic DNA extracted from soft organ tissues and bacteria
Achieve outstanding results when compared to other commercially available solutions
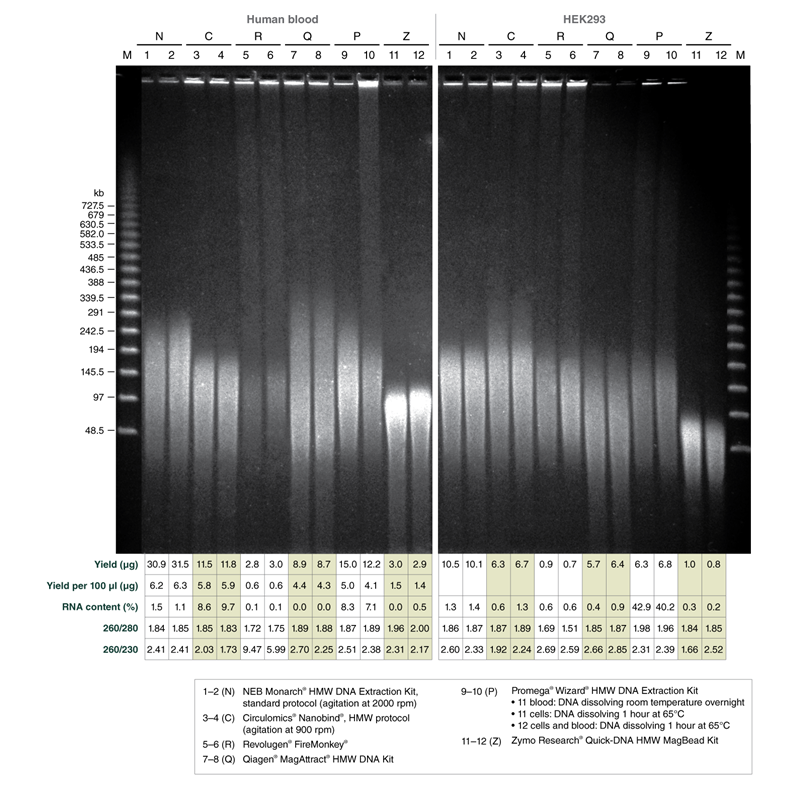
Monarch HMW DNA Extraction Kit produces high molecular weight DNA with excellent yields, purity, and fragment length as compared to other commercially available kits

Monarch samples (lanes 1-2) were purified at maximum agitation speed during lysis (2000 rpm). Variation in fragment length of cellular DNA using the standard protocols for Monarch and Circulomics (lanes 1-2 and 3-4, respectively) results from agitation speeds during lysis (Monarch: 2000 rpm, Circulomics: 900 rpm). All other data presented are duplicate samples from each different kit and the standard protocols dedicated to blood or cells were followed. Qiagen does not provide a protocol for cultured cells; a modified version of the blood protocol was followed. Samples were eluted in 100 μl, except for Zymo which was eluted in 50 μl according to their recommendations.
Yield and purity of the standard samples were analyzed on Trinean Dropsense 16 spectrophotometer (now Unchained Labs Lunatic. Reported blood sample yields were normalized per 100 μl. RNA content was determined by HPLC analysis of nucleoside content after digestion of 1 μg of eluted DNA with the Nucleoside Digestion Mix (NEB #M0649) . The optional RNase treatment was performed with the Zymo prep.
Excellent performance in long read sequencing
Single Run Sequencing Results of HEK293, Human Blood, and Mouse Kidney Samples on the Oxford Nanopore Technologies Platform
| HEK293 SAMPLE 1 | HEK293 SAMPLE 2 | HUMAN BLOOD SAMPLE 1 | HUMAN BLOOD SAMPLE 2 | MOUSE KIDNEY SAMPLE | |
|---|---|---|---|---|---|
| Mean read length (bases) | 21338.9 | 19249.9 | 21522.6 | 24677.7 | 27120.7 |
| Mean read quality | 12.8 | 13.2 | 13.4 | 13.3 | 13 |
| Median read length | 10388 | 9702 | 10130 | 12593 | 23150 |
| Median read quality | 13.2 | 13.7 | 13.9 | 13.8 | 13.5 |
| Number of reads | 377687 | 633636 | 538090 | 327314 | 164000 |
| Read length N50 (bases) | 45432 | 40415 | 46542 | 51394 | 44631 |
| Total bases | 8059414490 (8.1 Gb) |
12197410796 (12.2 Gb) |
11581090785 (11.6 Gb) |
8077351338 |
4447789727 (4.4 Gb) |
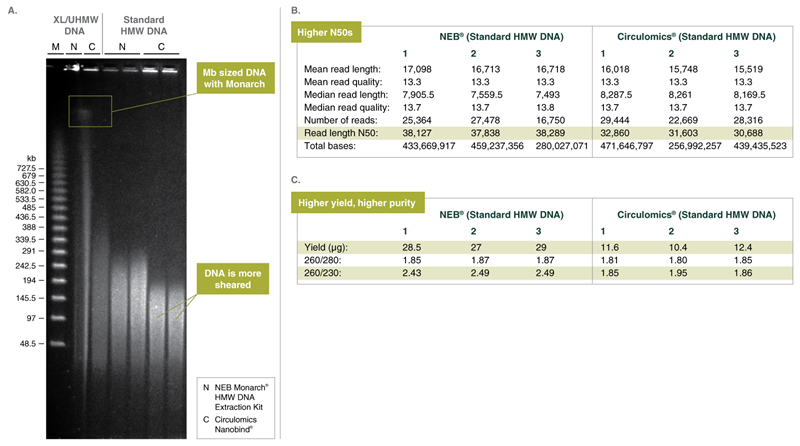
Monarch can purify Mb sized DNA from blood and provides excellent nanopore sequencing results
PacBio® Sequel I sequencing data from Monarch purified HMW DNA from human blood sheared to 75 kb (size selected to >30 kb)
Product Information
| Monarch® HMW DNA Extraction Kit for Cells & Blood | Cat No. | Concentration | SIze |
|---|---|---|---|
| T3050S | Not Applicable | 5 preps | |
| T3050L | Not Applicable | 50 preps | |
| Monarch® HMW DNA Extraction Kit for Tissue | Cat No. | Concentration | SIze |
| T3060S | Not Applicable | 5 preps | |
| T3060L | Not Applicable | 50 preps |