Pulse Chase
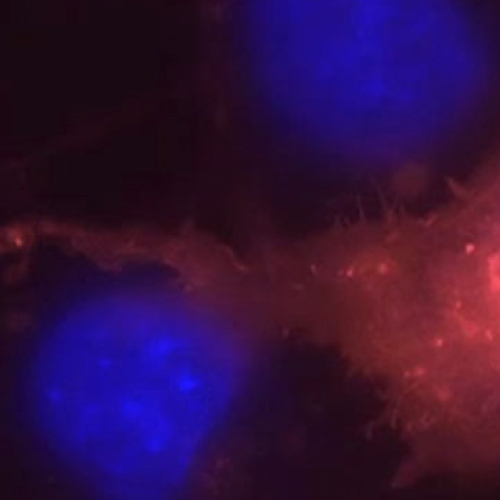
Pulse-chase experiments use labeled compounds to follow the dynamics of cellular processes and pathways. Molecules in a cell are continually being synthesized and degraded at various rates. Changes in molecule localizations and ____expression____ levels over time, can be detected by first “pulsing” or exposing cells to a labeled compound, then sequentially exposing the cells to the same compound, unlabeled, which is the “chase”. Compounds are commonly labeled with radioisotopes or fluorescent dyes. Turnover can be detected visually by microscopy techniques.
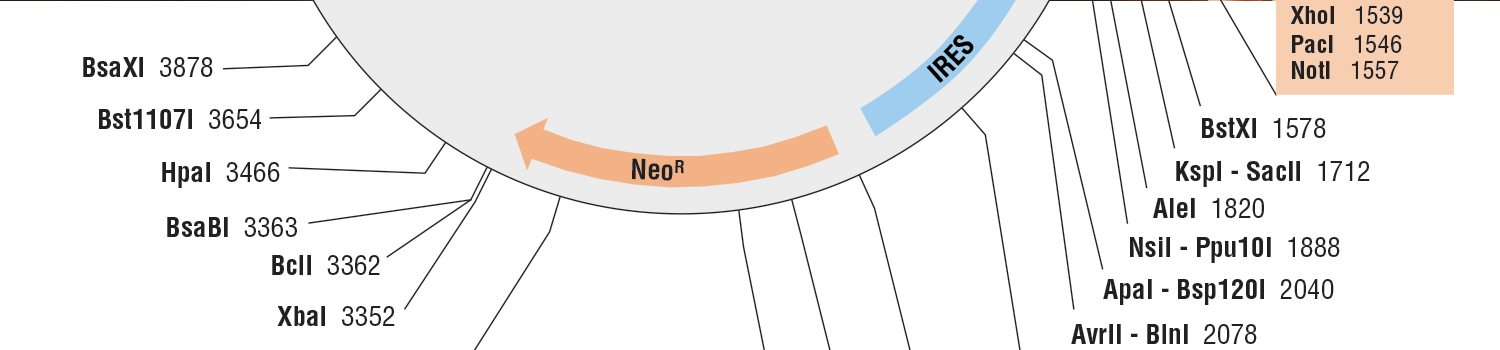
Various protein labeling methods for pulse-chase have specific application potentials. Reporter fluorescence fusion proteins are often used, but they can disrupt the native target protein's complexing behavior, be toxic to the cells, or affect target stability for pulse chase experiments. Fluorescence reporter fusion proteins also cannot be quenched. SNAP-tag® and CLIP-tag™ are useful for multicolor pulse-chase experiments. Sequential labeling with two or more fluorophores is possible. The non-overlapping substrate specificity of SNAP-tag and CLIP-tag permit simultaneous pulse-chase experiments to visualize different generations of two different proteins in one cell, further increasing the potential of the approach (1). Unlike with reporter fluorescence fusion proteins, like GFP, labeling of newly synthesized proteins can also be turned off using available blocking reagents (e.g., SNAP-Cell® Block). For development of pulse-chase techniques in thick tissues or animals (2), TIME-stamp tagging has advantages. Scalable proteomic analysis, detected by mass spectrometry, has been developed using a azidohomoalanine (azhal) label for rapid pulse-chase (3).
Feature
- 1
Clone and express once, then use with a variety of substrates
- 2
Non-toxic to living cells
- 3
Wide selection of fluorescent substrates
- 4
Highly specific covalent labeling
- 5
Simultaneous dual labeling
Product Information
Protein Localization
| Cat No. | SIze | |
|---|---|---|
| CLIP-Cell™ 505 | S9217S | 50 nmol |
| CLIP-Cell™ Block | S9220S | 100 nmol |
| CLIP-Cell™ TMR-Star | S9219S | 30 nmol |
| CLIP-Surface™ 488 | S9232S | 50 nmol |
| CLIP-Surface™ 547 | S9233S | 50 nmol |
| CLIP-Surface™ 647 | S9234S | 50 nmol |
| pCLIPf Vector | N9215S | 20 µg |
| pSNAPf Vector | N9183S | 20 µg |
| pSNAP-tag® (T7)-2 Vector | N9181S | 20 µg |
| SNAP-Biotin® | S9110S | 50 nmol |
| SNAP-Cell® 505-Star | S9103S | 50 nmol |
| SNAP-Cell® Block | S9106S | 100 nmol |
| SNAP-Cell® TMR-Star | S9105S | 30 nmol |
| SNAP-Cell® 430 | S9109S | 50 nmol |
| SNAP-Cell® 647-SiR | S9102S | 30 nmol |
| SNAP-Surface® 488 | S9124S | 50 nmol |
| SNAP-Surface® 549 | S9112S | 50 nmol |
| SNAP-Surface® 594 | S9134S | 50 nmol |
| SNAP-Surface® 649 | S9594S | 50 nmol |
| SNAP-Surface® Alexa Fluor® 488 | S9129S | 50 nmol |
| SNAP-Surface® Alexa Fluor® 546 | S9132S | 50 nmol |
| SNAP-Surface® Alexa Fluor® 647 | S9136S | 50 nmol |
| SNAP-Surface® Block | S9143S | 200 nmol |